A Precision Tool for Manipulating Mitochondrial DNA
Many mitochondrial diseases have been difficult to study and treat due to inherent challenges in accessing mitochondrial DNA (mtDNA). Now, researchers in Japan have optimized mitochondria-targeted compounds that can selectively alter the ratio of normal to mutant mtDNA in patient-derived stem cells. This technology enables the creation of research models with varying mutation loads and shows potential as a therapeutic strategy to reduce mutant mtDNA in patients, offering hope for the treatment of mitochondrial diseases.
Mitochondrial diseases affect approximately 1 in 5,000 people worldwide, causing debilitating symptoms ranging from muscle weakness to stroke-like episodes. Some of these conditions result from mutations in mitochondrial DNA, the genetic material present in these organelles. For patients with the common m.3243A>G mutation — which can cause MELAS syndrome (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes), as well as diabetes mellitus — treatments are still limited.
One of the main challenges in mitochondrial disease research is that patients often carry a mixture of normal and mutant mtDNA in their cells. This condition, known as heteroplasmy, hinders the development of targeted therapies, as the ratio of normal to mutant mtDNA can vary significantly from one tissue to another.
In addition, current basic research on mtDNA mutations faces major obstacles due to the lack of disease models. The complex relationship between mutation load (the percentage of mutant mtDNA) and disease severity remains poorly understood, in part because there are no tools available to precisely manipulate heteroplasmy levels in both directions. Without the ability to create cell models with varying mutation burdens, scientists are unable to effectively study how different proportions of mutant mtDNA relate to disease manifestation.
In response to this challenge, a research team led by Senior Assistant Professor Naoki Yahata from the Department of Developmental Biology at the Fujita Health University School of Medicine in Japan developed a technology capable of modifying heteroplasmy levels in cultured cells carrying the m.3243A>G mutation. The article was made available online on March 20, 2025, and will be published in the June 10, 2025 issue (Volume 36, Issue 2) of Molecular Therapy Nucleic Acids. The work was co-authored by Dr. Yu-ichi Goto from the National Center of Neurology and Psychiatry and Dr. Ryuji Hata from the Osaka Prefectural Hospital Organization.
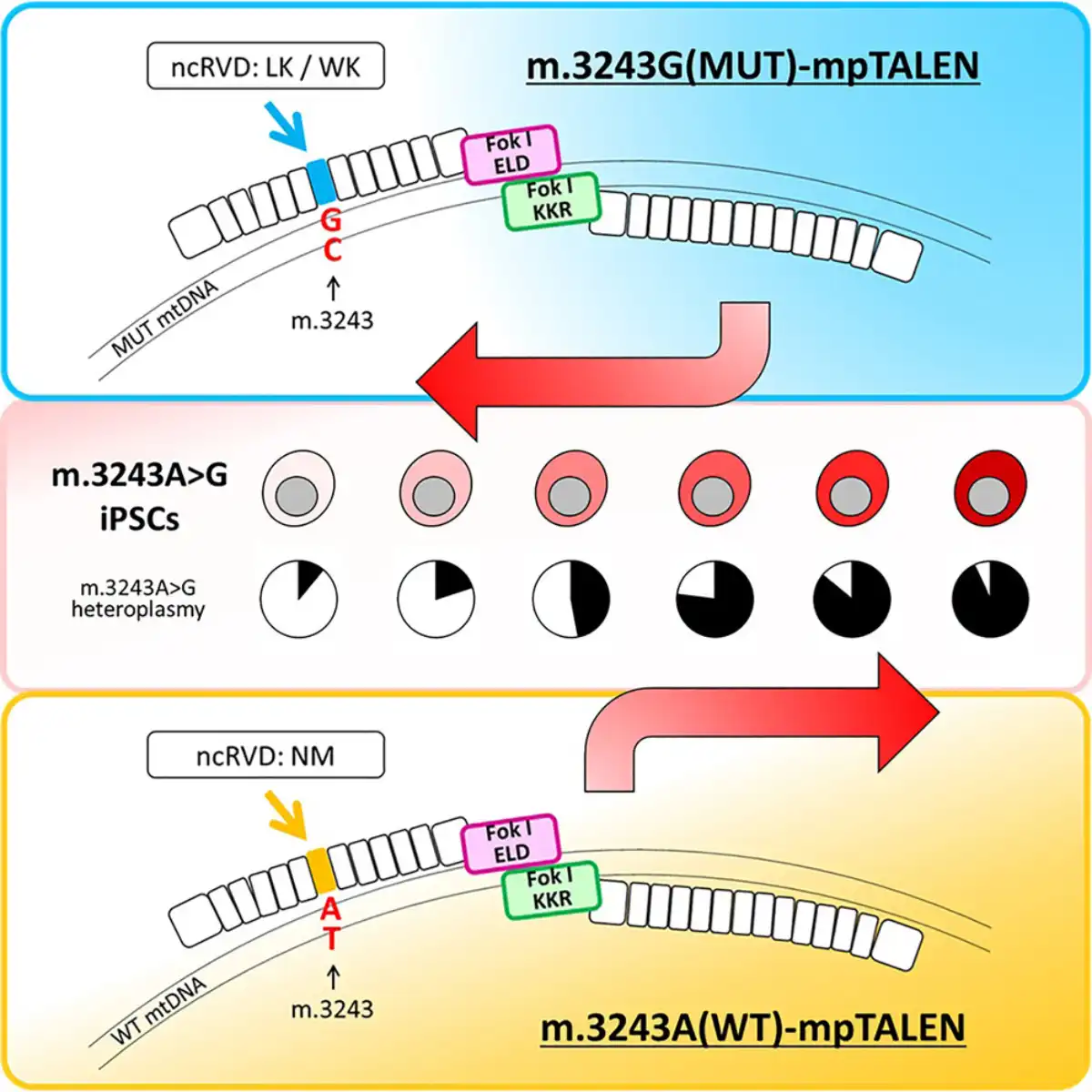
In the paper, the authors describe the development and application of platinum-conjugated mitochondria-targeted transcription activator-like effector nucleases (mpTALENs) — specialized enzymes that can selectively target and cleave specific DNA sequences. The researchers first established cultures of induced pluripotent stem cells (iPSCs) derived from patients carrying the m.3243A>G mutation and then designed two versions of the mpTALEN system: one to destroy the mutant mtDNA and another to destroy the normal mtDNA. This bidirectional approach enabled the generation of cells with mutation loads ranging from just 11% to as high as 97%, while preserving the cells’ ability to differentiate into various tissue types. “Our study is the first to demonstrate an increase in the proportion of pathogenic mtDNA using programmable nucleases,” notes Dr. Yahata.
The main innovations of the approach included the use of novel unconventional variable dipeptide residues and obligate heterodimeric FokI nuclease domains, which increased the specificity of the technology and reduced off-target degradation of mtDNA. The team also employed additional techniques, such as uridine supplementation, to establish stable cell lines with different mutation loads, including those that would normally have growth disadvantages. “Our results demonstrate that the optimization process of mpTALENs has created a useful tool for altering heteroplasmy levels in iPSCs with the m.3243A>G mutation, expanding their potential for studying the pathology of these mutations. This enhanced efficiency also holds promise for the use of mpTALENs in therapeutic strategies to treat patients with mitochondrial diseases related to the m.3243A>G mutation,” Yahata stated.
- See also: How aging affects bone cells
Overall, the study represents a significant advancement in mitochondrial medicine for several reasons. First, it provides researchers with multiple isogenic cell lines — genetically identical to each other — that differ only in their level of heteroplasmy, enabling precise study of how mutation load affects disease manifestation. Second, it suggests that mpTALEN technology may have future therapeutic value by enabling the reduction of mutant mtDNA burden in patients.
“Our proposed method can be adapted for other mtDNA mutations and contribute to understanding their respective pathologies, as well as facilitate the development of new treatments, potentially benefiting patients with various forms of mitochondrial diseases,” concludes Dr. Yahata.